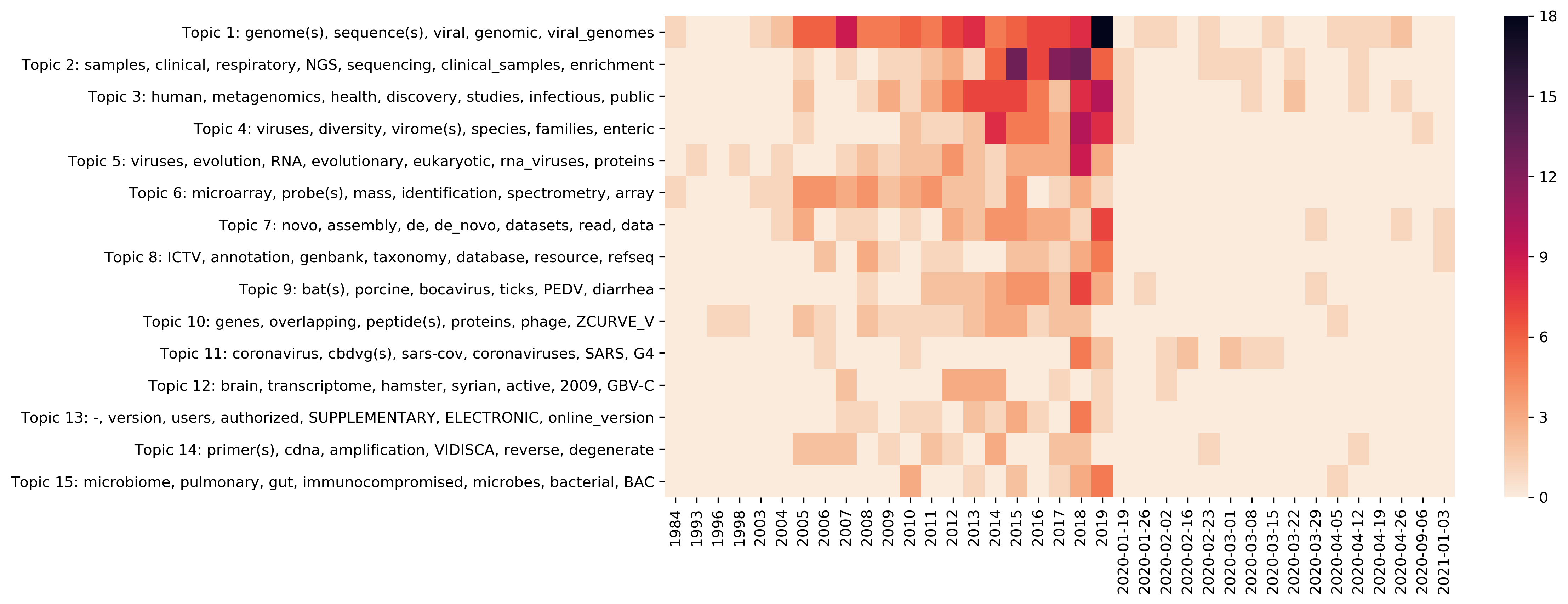
| t01 |
genome(s), sequence(s), viral, genomic, viral_genomes, RNA, virus, based |
121 |
5 |
| t02 |
sample(s), clinical, respiratory, NGS, sequencing, clinical_samples, enrichment, detection, sensitivity |
73 |
1 |
| t03 |
human, metagenomics, health, discovery, studies, infectious, public, approaches, infectious_diseases, disease |
67 |
3 |
| t04 |
viruses, diversity, virome(s), species, families, enteric, picornaviridae, associated_with, viral_diversity |
48 |
0 |
| t05 |
viruses, evolution, RNA, evolutionary, eukaryotic, rna_viruses, protein(s), classes, virus |
39 |
0 |
| t06 |
microarray(s), probe(s), mass, identification, spectrometry, array, methods, MS |
41 |
0 |
| t07 |
novo, assembly, de, de_novo, datasets, read, data, short, contig, problem |
37 |
0 |
| t08 |
ICTV, annotation, genbank, taxonomy, database, resource, refseq, taxonomic, virus_taxonomy, submissions |
22 |
0 |
| t09 |
bat(s), porcine, bocavirus, ticks, PEDV, diarrhea, USA, circovirus, named |
32 |
0 |
| t10 |
genes, overlapping, peptide(s), proteins, phage, ZCURVE_V, codon, usage, archaeal |
25 |
1 |
| t11 |
coronavirus, cbdvg(s), sars-cov, coronaviruses, SARS, G4, v2.0, RSV, position |
16 |
7 |
| t12 |
brain, transcriptome, hamster, syrian, active, 2009, GBV-C, H., H1N1, brain_specimens |
14 |
0 |
| t13 |
-, version, users, authorized, SUPPLEMENTARY, ELECTRONIC, online_version, MATERIAL, online, supplementary |
17 |
0 |
| t14 |
primer(s), cdna, amplification, VIDISCA, reverse, degenerate, hexanucleotides, transcription, ribosomal |
19 |
1 |
| t15 |
microbiome, pulmonary, gut, immunocompromised, microbes, bacterial, BAC, giant, manipulation, microbiota |
16 |
0 |