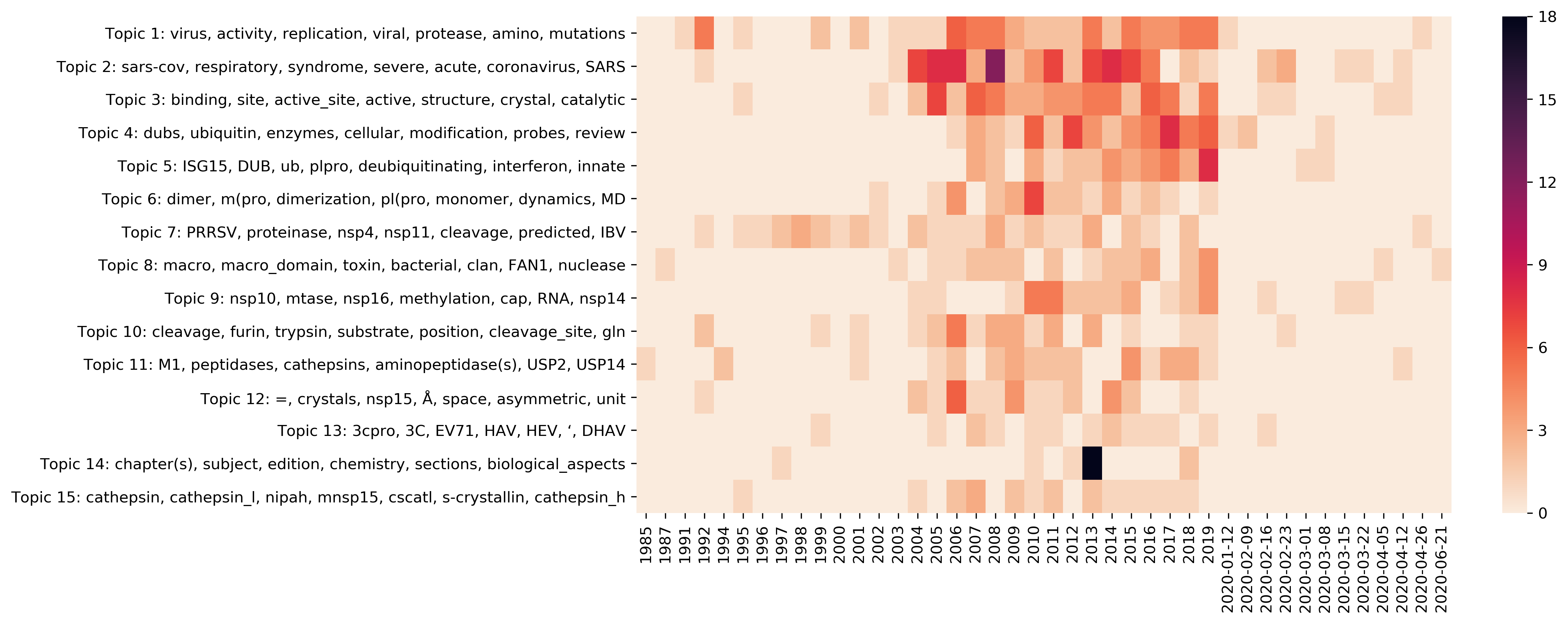
| t01 |
virus, activity, replication, viral, protease, amino, mutations, nsp2, RNA, in_vitro |
71 |
1 |
| t02 |
sars-cov, respiratory, syndrome, severe, acute, coronavirus, SARS, 3clpro, inhibitors, protease |
93 |
5 |
| t03 |
binding, site, active_site, active, structure(s), crystal, catalytic, enzyme, complex |
71 |
3 |
| t04 |
dubs, ubiquitin, enzymes, cellular, modification, probes, review, adp-ribosylation, proteins, processes |
60 |
0 |
| t05 |
ISG15, DUB, ub, plpro, deubiquitinating, interferon, innate, ubiquitin, immune, isgylation |
42 |
0 |
| t06 |
dimer, m(pro, dimerization, pl(pro, monomer, dynamics, MD, extra, dimeric, catalytic_machinery |
31 |
0 |
| t07 |
PRRSV, proteinase, nsp4, nsp11, cleavage, predicted, IBV, reproductive, MHV, 3c-like_proteinase |
36 |
0 |
| t08 |
macro, macro_domain, toxin, bacterial, clan, FAN1, nuclease, endou, superfamily, domains |
28 |
1 |
| t09 |
nsp10, mtase, nsp16, methylation, cap, RNA, nsp14, methyltransferase, mrna, polymerase |
32 |
1 |
| t10 |
cleavage, furin, trypsin, substrate, position(s), cleavage_site, gln, P3, peptide |
30 |
1 |
| t11 |
M1, peptidases, cathepsins, aminopeptidase(s), USP2, USP14, serine, enhance, tissue |
31 |
0 |
| t12 |
=, crystals, nsp15, Å, space, asymmetric, unit, space_group, parameters, resolution |
27 |
1 |
| t13 |
3cpro, 3C, EV71, HAV, HEV, ‘, DHAV, x-domain, 2A, dhav_3c_protease |
15 |
0 |
| t14 |
chapter(s), subject, edition, chemistry, sections, biological_aspects, preparation, cleave_proteins, third_edition |
23 |
0 |
| t15 |
cathepsin, cathepsin_l, nipah, mnsp15, cscatl, s-crystallin, cathepsin_h, cscatd, catalytic_site, WT |
19 |
0 |