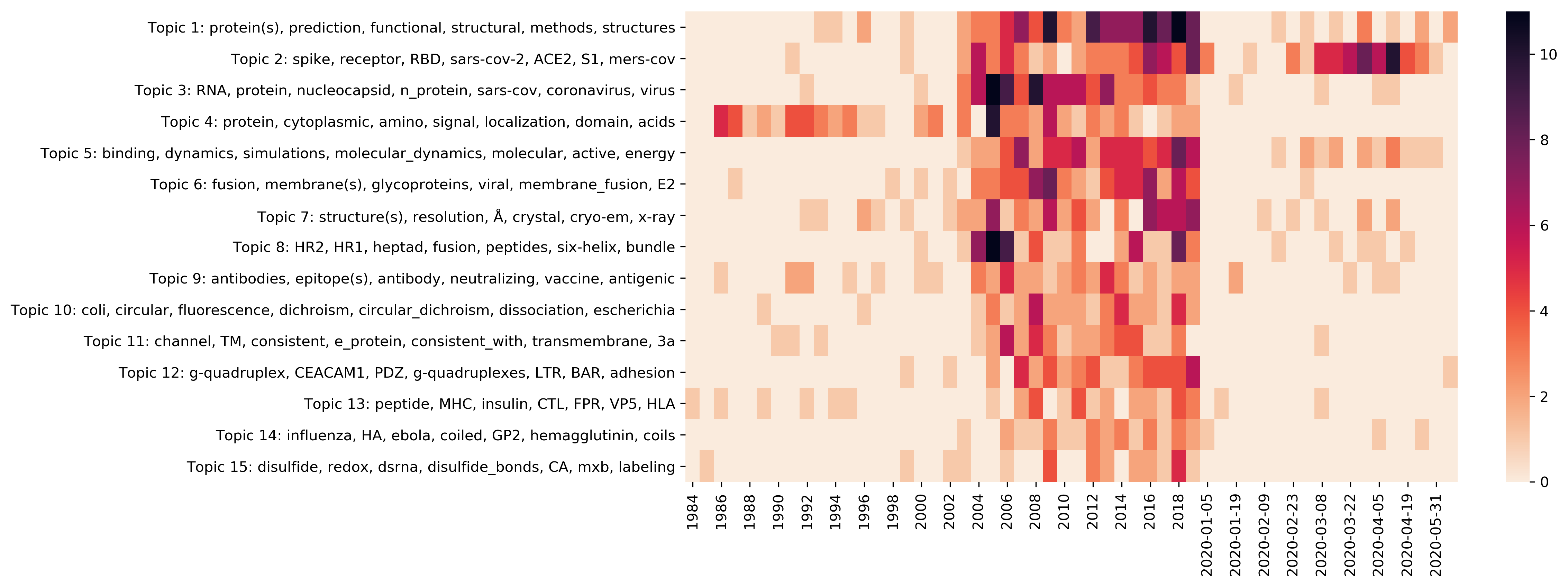
| t01 |
protein(s), prediction, functional, structural, method(s), structures, 3D, sequence |
122 |
5 |
| t02 |
spike, receptor, RBD, sars-cov-2, ACE2, S1, mers-cov, receptor-binding, host, coronavirus |
120 |
53 |
| t03 |
RNA, protein, nucleocapsid, n_protein, sars-cov, coronavirus, virus, interaction, acute, syndrome |
95 |
5 |
| t04 |
protein, cytoplasmic, amino, signal, localization, domain, acids, fragments, membrane, amino_acids |
80 |
1 |
| t05 |
binding, dynamics, simulations, molecular_dynamics, molecular, active, energy, docking, catalytic, free |
89 |
12 |
| t06 |
fusion, membrane(s), glycoprotein(s), viral, membrane_fusion, E2, HCV, lipid |
73 |
0 |
| t07 |
structure(s), resolution, Å, crystal, cryo-em, x-ray, complex, crystal_structure, solved |
74 |
0 |
| t08 |
HR2, HR1, heptad, fusion, peptides, six-helix, bundle, repeat, HR, inhibitors |
65 |
6 |
| t09 |
antibodies, epitope(s), antibody, neutralizing, vaccine, antigenic, monoclonal, FIPV, neutralizing_antibodies |
52 |
3 |
| t10 |
coli, circular, fluorescence, dichroism, circular_dichroism, dissociation, escherichia, purification, unfolding, escherichia_coli |
42 |
0 |
| t11 |
channel, TM, consistent, e_protein, consistent_with, transmembrane, 3a, lipid, ion, orientation |
44 |
0 |
| t12 |
g-quadruplex, CEACAM1, PDZ, g-quadruplexes, LTR, BAR, adhesion, A3, domains, neck |
48 |
0 |
| t13 |
peptide, MHC, insulin, CTL, FPR, VP5, HLA, alleles, presentation, t-cell |
35 |
1 |
| t14 |
influenza, HA, ebola, coiled, GP2, hemagglutinin, coils, gb, ebola_virus, NA |
31 |
0 |
| t15 |
disulfide, redox, dsrna, disulfide_bonds, CA, mxb, labeling, IL-2, σ1, nanoparticles |
25 |
0 |