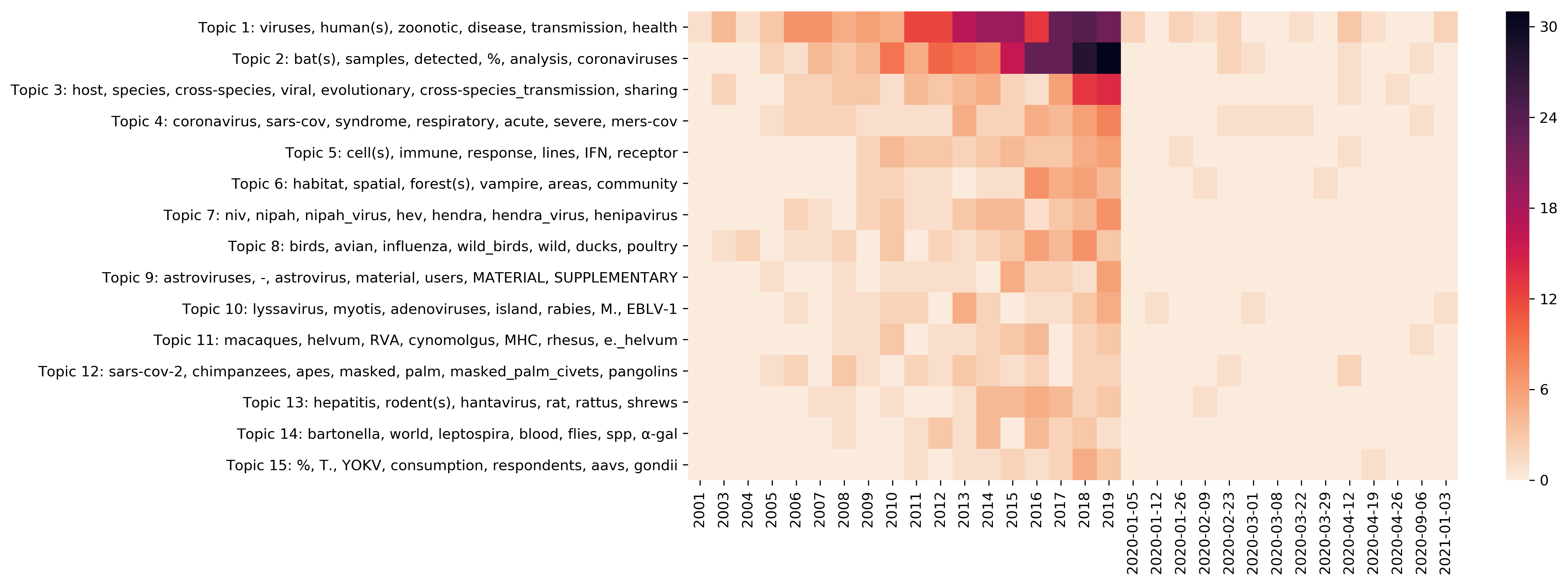
| t01 |
viruses, human(s), zoonotic, disease(s), transmission, health, animal(s) |
214 |
10 |
| t02 |
bat(s), samples, detected, %, analysis, coronaviruses, sequences, species, phylogenetic |
181 |
6 |
| t03 |
host, species, cross-species, viral, evolutionary, cross-species_transmission, sharing, adaptation, pathogen, shifts |
67 |
2 |
| t04 |
coronavirus, sars-cov, syndrome, respiratory, acute, severe, mers-cov, SARS, acute_respiratory_syndrome, middle |
48 |
9 |
| t05 |
cell(s), immune, response, lines, IFN, receptor, expression, replication, antiviral |
40 |
0 |
| t06 |
habitat, spatial, forest(s), vampire, areas, community, urban, dynamics, abundance |
32 |
0 |
| t07 |
niv, nipah, nipah_virus, hev, hendra, hendra_virus, henipavirus, paramyxoviridae, sera, antibodies |
36 |
0 |
| t08 |
bird(s), avian, influenza, wild_birds, wild, duck(s), poultry, anas |
38 |
1 |
| t09 |
astroviruses, -, astrovirus, material, users, MATERIAL, SUPPLEMENTARY, authorized, ELECTRONIC, supplementary |
22 |
0 |
| t10 |
lyssavirus, myotis, adenoviruses, island, rabies, M., EBLV-1, daubentonii, free-tailed, colonies |
27 |
1 |
| t11 |
macaque(s), helvum, RVA, cynomolgus, MHC, rhesus, e._helvum, eidolon_helvum, E. |
22 |
0 |
| t12 |
sars-cov-2, chimpanzees, apes, masked, palm, masked_palm_civets, pangolins, simian, gorillas, retroviruses |
25 |
2 |
| t13 |
hepatitis, rodent(s), hantavirus, rat, rattus, shrews, HCV, norvegicus, hantaviruses |
27 |
0 |
| t14 |
bartonella, world, leptospira, blood, flies, spp, α-gal, parasites, =, α-gal_epitopes |
20 |
0 |
| t15 |
%, T., YOKV, consumption, respondents, aavs, gondii, foxes, letea, wild_pigs |
17 |
0 |